missing translation for 'onlineSavingsMsg'
Learn More
Learn More
G3BP1 Antibody, Novus Biologicals™
Rabbit Polyclonal Antibody has been used in 1 publication
Brand: Novus Biologicals NBP1-83404-25ul
This item is not returnable.
View return policy
Description
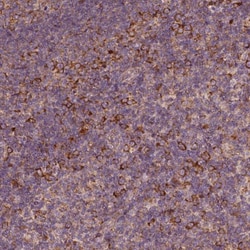
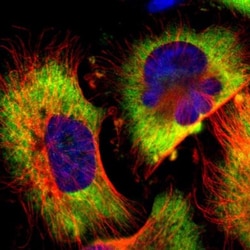
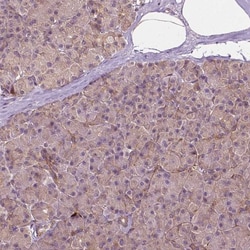
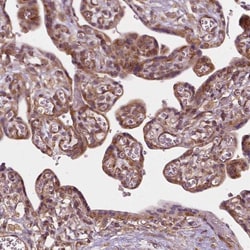
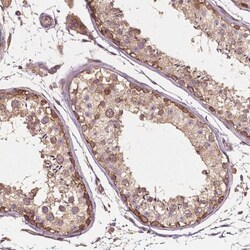
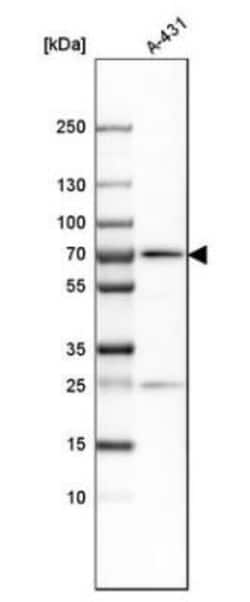
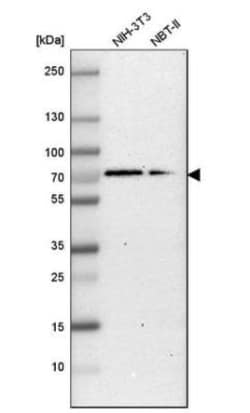
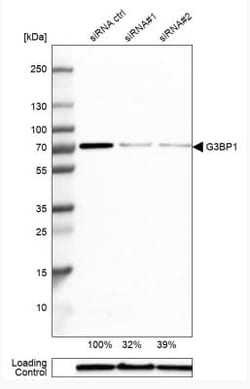
G3BP1 Polyclonal specifically detects G3BP1 in Human, Mouse, Rat samples. It is validated for Western Blot, Immunohistochemistry, Immunocytochemistry/Immunofluorescence, Immunohistochemistry-Paraffin, Knockdown Validated.
Specifications
| G3BP1 | |
| Polyclonal | |
| Western Blot 0.04-0.4 ug/ml, Immunohistochemistry 1:50 - 1:200, Immunocytochemistry/Immunofluorescence 0.25-2 ug/ml, Immunohistochemistry-Paraffin 1:50 - 1:200, Knockdown Validated | |
| ATP-dependent DNA helicase VIII, EC 3.6.1, EC 3.6.4.12, EC 3.6.4.13, G3BP-1, G3BPRas-GTPase-activating protein SH3-domain-binding protein, GAP binding protein, GAP SH3 domain-binding protein 1, GTPase activating protein (SH3 domain) binding protein 1, hDH VIII, HDH-VIII, MGC111040, ras GTPase-activating protein-binding protein 1, RasGAP-associated endoribonuclease G3BP | |
| Rabbit | |
| Affinity Purified | |
| RUO | |
| Primary | |
| Specificity of human G3BP1 antibody verified on a Protein Array containing target protein plus 383 other non-specific proteins. | |
| Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. |
| Western Blot, Immunohistochemistry, Immunocytochemistry, Immunofluorescence, Immunohistochemistry (Paraffin) | |
| Unconjugated | |
| PBS (pH 7.2) and 40% Glycerol with 0.02% Sodium Azide | |
| G3BP1 | |
| This antibody was developed against Recombinant Protein corresponding to amino acids:FRYQDEVFGGFVTEPQEESEEEVEEPEERQQTPEVVPDDSGTFYDQAVVSNDMEEHLEEPVAEPEPDPEPEPEQEPVSEIQEEKPEPVLEETAP | |
| 25 μL | |
| Core ESC Like Genes, Stem Cell Markers | |
| 10146 | |
| Human, Mouse, Rat | |
| IgG |
Product Content Correction
Your input is important to us. Please complete this form to provide feedback related to the content on this product.
Product Title
For Research Use Only
Spot an opportunity for improvement?Share a Content Correction